Wrangling
R for Data Science
Basel R Bootcamp

Basel R Bootcamp

Overview
In this practical you’ll practice “data wrangling” with the dplyr and tidyr packages (part of the tidyverse collection of packages).
By the end of this practical you will know how to:
- Change column names, select specific columns
- Create new columns
- Filter rows of data based on multiple criteria
- Combine (aka, ‘join’) multiple data sets through key columns
- Convert data between wide and long formats
Tasks
A - Setup
- Open your
BaselRBootcampR project. It should already have the folders1_Dataand2_Code. Make sure that the data files listed in theDatasetssection above are in your1_Datafolder
# Done!- Open a new R script. At the top of the script, using comments, write your name and the date. Save it as a new file called
wrangling_practical.Rin the2_Codefolder.
# Done!- Using
library()load thetidyversepackage (if you don’t have it, you’ll need to install it withinstall.packages())!
# Load packages necessary for this script
library(tidyverse)For this practical, we’ll use the trial_act data, this is the result of a randomized clinical trial comparing the effects of different medications on adults infected with the human immunodeficiency virus (You can learn more about the trial here).
- Using the following template, load the
trial_act.csvdata into R and store it as a new object calledtrial_act(Hint: Don’t type the path directly! Use the “tab” completion!).
# Load trial_act.csv from the data folder in your
# working directory as a new object called trial_act
trial_act <- read_csv(file = "XX/XX")trial_act <- read_csv(file = "1_Data/trial_act.csv")- Using the same code structure, load the
trial_act_demo_fake.csvdata as a new dataframe calledtrial_act_demo.
# Load trial_act_demo_fake.csv from the data folder in your
# working directory as a new object called trial_act_demo
XX <- XX(XX = "XX/XX")trial_act_demo <- read_csv(file = "1_Data/trial_act_demo_fake.csv")- Take a look at the first few rows of the datasets by printing them to the console.
# Print trial_act object
trial_act# A tibble: 2,139 x 27
pidnum age wtkg hemo homo drugs karnof oprior z30 zprior preanti
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 10056 48 89.8 0 0 0 100 0 0 1 0
2 10059 61 49.4 0 0 0 90 0 1 1 895
3 10089 45 88.5 0 1 1 90 0 1 1 707
4 10093 47 85.3 0 1 0 100 0 1 1 1399
5 10124 43 66.7 0 1 0 100 0 1 1 1352
6 10140 46 88.9 0 1 1 100 0 1 1 1181
7 10165 31 73.0 0 1 0 100 0 1 1 930
8 10190 41 66.2 0 1 1 100 0 1 1 1329
9 10198 40 82.6 0 1 0 90 0 1 1 1074
10 10229 35 78.0 0 1 0 100 0 1 1 964
# … with 2,129 more rows, and 16 more variables: race <dbl>, gender <dbl>,
# str2 <dbl>, strat <dbl>, symptom <dbl>, treat <dbl>, offtrt <dbl>,
# cd40 <dbl>, cd420 <dbl>, cd496 <dbl>, r <dbl>, cd80 <dbl>,
# cd820 <dbl>, cens <dbl>, days <dbl>, arms <dbl># Print trial_act_demo object
trial_act_demo# A tibble: 2,139 x 3
pidnum exercise education
<dbl> <dbl> <chr>
1 10931 4 HS
2 11971 1 <HS
3 330244 1 HS
4 270879 1 BA
5 11435 0 <HS
6 270849 0 PHD
7 10840 1 <HS
8 241099 3 <HS
9 71371 2 BA
10 241370 0 MS
# … with 2,129 more rows- Use the the
summary()function to print more details on the columns of the datasets.
summary(trial_act) pidnum age wtkg hemo
Min. : 10056 Min. :12.0 Min. : 31.0 Min. :0.000
1st Qu.: 81446 1st Qu.:29.0 1st Qu.: 66.7 1st Qu.:0.000
Median :190566 Median :34.0 Median : 74.4 Median :0.000
Mean :248778 Mean :35.2 Mean : 75.1 Mean :0.084
3rd Qu.:280277 3rd Qu.:40.0 3rd Qu.: 82.6 3rd Qu.:0.000
Max. :990077 Max. :70.0 Max. :159.9 Max. :1.000
homo drugs karnof oprior
Min. :0.000 Min. :0.000 Min. : 70.0 Min. :0.000
1st Qu.:0.000 1st Qu.:0.000 1st Qu.: 90.0 1st Qu.:0.000
Median :1.000 Median :0.000 Median :100.0 Median :0.000
Mean :0.661 Mean :0.131 Mean : 95.4 Mean :0.022
3rd Qu.:1.000 3rd Qu.:0.000 3rd Qu.:100.0 3rd Qu.:0.000
Max. :1.000 Max. :1.000 Max. :100.0 Max. :1.000
z30 zprior preanti race gender
Min. :0.00 Min. :1 Min. : 0 Min. :0.000 Min. :0.000
1st Qu.:0.00 1st Qu.:1 1st Qu.: 0 1st Qu.:0.000 1st Qu.:1.000
Median :1.00 Median :1 Median : 142 Median :0.000 Median :1.000
Mean :0.55 Mean :1 Mean : 379 Mean :0.288 Mean :0.828
3rd Qu.:1.00 3rd Qu.:1 3rd Qu.: 740 3rd Qu.:1.000 3rd Qu.:1.000
Max. :1.00 Max. :1 Max. :2851 Max. :1.000 Max. :1.000
str2 strat symptom treat
Min. :0.000 Min. :1.00 Min. :0.000 Min. :0.000
1st Qu.:0.000 1st Qu.:1.00 1st Qu.:0.000 1st Qu.:1.000
Median :1.000 Median :2.00 Median :0.000 Median :1.000
Mean :0.586 Mean :1.98 Mean :0.173 Mean :0.751
3rd Qu.:1.000 3rd Qu.:3.00 3rd Qu.:0.000 3rd Qu.:1.000
Max. :1.000 Max. :3.00 Max. :1.000 Max. :1.000
offtrt cd40 cd420 cd496
Min. :0.000 Min. : 0 Min. : 49 Min. : 0
1st Qu.:0.000 1st Qu.: 264 1st Qu.: 269 1st Qu.: 209
Median :0.000 Median : 340 Median : 353 Median : 321
Mean :0.363 Mean : 351 Mean : 371 Mean : 329
3rd Qu.:1.000 3rd Qu.: 423 3rd Qu.: 460 3rd Qu.: 440
Max. :1.000 Max. :1199 Max. :1119 Max. :1190
NA's :797
r cd80 cd820 cens
Min. :0.000 Min. : 40 Min. : 124 Min. :0.000
1st Qu.:0.000 1st Qu.: 654 1st Qu.: 632 1st Qu.:0.000
Median :1.000 Median : 893 Median : 865 Median :0.000
Mean :0.627 Mean : 987 Mean : 935 Mean :0.244
3rd Qu.:1.000 3rd Qu.:1207 3rd Qu.:1146 3rd Qu.:0.000
Max. :1.000 Max. :5011 Max. :6035 Max. :1.000
days arms
Min. : 14 Min. :0.00
1st Qu.: 727 1st Qu.:1.00
Median : 997 Median :2.00
Mean : 879 Mean :1.52
3rd Qu.:1091 3rd Qu.:3.00
Max. :1231 Max. :3.00
summary(trial_act_demo) pidnum exercise education
Min. : 10056 Min. :0.0 Length:2139
1st Qu.: 81446 1st Qu.:1.0 Class :character
Median :190566 Median :1.0 Mode :character
Mean :248778 Mean :2.2
3rd Qu.:280277 3rd Qu.:3.0
Max. :990077 Max. :7.0 - Use the
View()function to view the entire dataframes in new windows
View(trial_act)
View(trial_act_demo)B - Change column names with rename()
- Print the names of the
trial_actdata withnames(XXX)
names(XXX)names(trial_act) [1] "pidnum" "age" "wtkg" "hemo" "homo" "drugs" "karnof"
[8] "oprior" "z30" "zprior" "preanti" "race" "gender" "str2"
[15] "strat" "symptom" "treat" "offtrt" "cd40" "cd420" "cd496"
[22] "r" "cd80" "cd820" "cens" "days" "arms" - One of the columns is currently called
wtkg. Usingrename(), change the name of this column toweight_kg. Be sure to assign the result back totrial_actto change it!
# Change the name to weight_kg from wtkg
trial_act <- trial_act %>%
rename(XX = XX) # NEW = OLDtrial_act <- trial_act %>%
rename(weight_kg = wtkg)- Look at the names of your
trial_actdataframe again usingnames(), do you now see the columnweight_kg?
names(trial_act) [1] "pidnum" "age" "weight_kg" "hemo" "homo"
[6] "drugs" "karnof" "oprior" "z30" "zprior"
[11] "preanti" "race" "gender" "str2" "strat"
[16] "symptom" "treat" "offtrt" "cd40" "cd420"
[21] "cd496" "r" "cd80" "cd820" "cens"
[26] "days" "arms" - One of the columns is called
age, change it toage_y(to specify that age is in years).
trial_act <- trial_act %>%
rename(age_y = age)C - Select columns with select()
- Using the
select()function, select only the columnage_yand print the result (but don’t assign it to anything). Do you see only theage_ycolumn now?
XX %>%
select(XX)trial_act %>%
select(age_y)# A tibble: 2,139 x 1
age_y
<dbl>
1 48
2 61
3 45
4 47
5 43
6 46
7 31
8 41
9 40
10 35
# … with 2,129 more rows- Using
select()select the columnspidnum,age_y,gender, andweight_kg(but don’t assign the result to anything)
XX %>%
select(XX, XX, XX, XX)trial_act %>%
select(pidnum, age_y, gender, weight_kg)# A tibble: 2,139 x 4
pidnum age_y gender weight_kg
<dbl> <dbl> <dbl> <dbl>
1 10056 48 0 89.8
2 10059 61 0 49.4
3 10089 45 1 88.5
4 10093 47 1 85.3
5 10124 43 1 66.7
6 10140 46 1 88.9
7 10165 31 1 73.0
8 10190 41 1 66.2
9 10198 40 1 82.6
10 10229 35 1 78.0
# … with 2,129 more rows- Imagine that a colleague of yours wants an anonymised dataframe called
trial_act_anon_that does not contain the columnspidnumandage_y. Create this dataframe by selecting all columns intrial_actexceptpidnumandage_y(hint: use the notationselect(-XXX, -XXX))to select everything except specified columns). Assign the result to a new object calledtrial_act_anon
XX <- XX %>%
select(-XX, -XX)- Create a new dataframe called
CD4_widewhich contains the following columns:pidnum,arms,cd40,cd420, andcd496. Thecd40,cd420, andcd496columns show patient’s CD4 T cell counts at baseline, 20 weeks, and 96 weeks. After you createCD4_wide, print it to make sure it worked!
XX <- trial_act %>%
select(XX, XX, XX, XX, XX)CD4_wide <- trial_act %>%
select(pidnum, arms, cd40, cd420, cd496)- Did you know you can easily select all columns that start with specific characters using
starts_with()? Try adapting the following code to get the same result you got before.
CD4_wide <- trial_act %>%
select(pidnum, arms, starts_with("XXX"))CD4_wide <- trial_act %>%
select(pidnum, arms, starts_with("cd"))D - Add new columns with mutate()
- Using the
mutate()function, add the columnage_mwhich shows each patient’s age in months instead of years (Hint: Just multiplyage_yby 12!)
trial_act <- trial_act %>%
mutate(XX = XX * 12)trial_act <- trial_act %>%
mutate(age_m = age_y * 12)- Using
mutate, add the following new columns totrial_act. (Try combining these into one call to themutate()function!)
weight_lb: Weight in lbs instead of kilograms. You can do this by multiplyingweight_kgby 2.2.cd_change_20: Change in CD4 T cell count from baseline to 20 weeks. You can do this by takingcd420 - cd40cd_change_960: Change in CD4 T cell count from baseline to 96 weeks. You can do this by takingcd496 - cd40
XXX <- XXX %>%
mutate(weight_lb = XXX,
cd_change_20 = XXX,
XXX = XXX)trial_act <- trial_act %>%
mutate(weight_lb = weight_kg * 2.2,
cd_change_20 = cd420 - cd40,
cd_change_960 = cd496 - cd40)- If you look at the
gendercolumn, you will see that it is numeric (0s and 1s). Using themutate()andcase_when()functions, create a new column calledgender_charwhich shows the gender as a string, where 0 = “female” and 1 = “male”:
# Create gender_char which shows gender as a string
trial_act <- trial_act %>%
mutate(
gender_char = case_when(
gender == XX ~ "XX",
gender == XX ~ "XX"))trial_act <- trial_act %>%
mutate(
gender_char = case_when(
gender == 0 ~ "female",
gender == 1 ~ "male"))- The column
armsis also numeric and not very meaningful. Create a new columnarms_charcontains the names of the arms. Here is a table of the mapping
| arms | arms_char |
|---|---|
| 0 | zidovudine |
| 1 | zidovudine and didanosine |
| 2 | zidovudine and zalcitabine |
| 3 | didanosine |
trial_act <- trial_act %>%
mutate(
arms_char = case_when(
arms == 0 ~ "zidovudine",
arms == 1 ~ "zidovudine and didanosine",
arms == 2 ~ "zidovudine and zalcitabine",
arms == 3 ~ "didanosine"))- If you haven’t already, try putting all the code for your previous questions in one call to
mutate(). That is, in one block of code, createage_m,weight_lb,cd_change_20,cd_change_960andgender_charandarms_charusing themutate()function only once. Here’s how your code should look:
trial_act <- trial_act %>%
mutate(
age_m = XXX,
weight_lb = XXX,
cd_change_20 = XXX,
cd_change_960 = XXX,
gender_char = XXX,
arms_char = XXX
)trial_act <- trial_act %>%
mutate(
age_m = age_y * 12,
weight_lb = weight_kg * 2.2,
cd_change_20 = cd420 - cd40,
cd_change_960 = cd496 - cd40,
gender_char = case_when(
gender == 0 ~ "female",
gender == 1 ~ "male"),
arms_char = case_when(
arms == 0 ~ "zidovudine",
arms == 1 ~ "zidovudine and didanosine",
arms == 2 ~ "zidovudine and zalcitabine",
arms == 3 ~ "didanosine")
)E - Arrange rows with arrange()
- Using the
arrange()function, arrange thetrial_actdata in ascending order ofage_y(from lowest to highest). After you do, print the data to make sure it worked!
trial_act <- trial_act %>%
arrange(XXX)trial_act <- trial_act %>%
arrange(age_y)
trial_act# A tibble: 2,139 x 33
pidnum age_y weight_kg hemo homo drugs karnof oprior z30 zprior
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 940533 12 41.4 1 0 0 100 0 0 1
2 950037 12 53.1 1 0 0 100 0 1 1
3 950056 12 31 1 0 0 100 0 1 1
4 910034 13 32.7 1 0 0 100 0 1 1
5 940534 13 62.9 1 0 0 100 0 0 1
6 960014 13 48.5 1 0 0 100 0 0 1
7 310767 14 65 1 0 0 100 0 1 1
8 920050 14 54.2 1 0 0 100 0 1 1
9 940544 14 41.1 1 0 0 100 0 0 1
10 950061 14 64.3 1 0 0 100 0 1 1
# … with 2,129 more rows, and 23 more variables: preanti <dbl>,
# race <dbl>, gender <dbl>, str2 <dbl>, strat <dbl>, symptom <dbl>,
# treat <dbl>, offtrt <dbl>, cd40 <dbl>, cd420 <dbl>, cd496 <dbl>,
# r <dbl>, cd80 <dbl>, cd820 <dbl>, cens <dbl>, days <dbl>, arms <dbl>,
# age_m <dbl>, weight_lb <dbl>, cd_change_20 <dbl>, cd_change_960 <dbl>,
# gender_char <chr>, arms_char <chr>- Now arrange the data in descending order of
age_y(from highest to lowest). After, look the data to make sure it worked. To arrange data in descending order, just includedesc()around the variable
trial_act <- trial_act %>%
arrange(desc(XXX))trial_act <- trial_act %>%
arrange(desc(age_y))
trial_act# A tibble: 2,139 x 33
pidnum age_y weight_kg hemo homo drugs karnof oprior z30 zprior
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 11438 70 73.9 0 1 0 100 0 1 1
2 211360 70 63.1 0 1 0 90 0 0 1
3 211284 69 81.6 0 1 0 100 0 0 1
4 50580 68 90.5 0 1 1 100 0 1 1
5 81127 68 70.8 0 1 0 90 0 1 1
6 10924 67 71 0 1 0 100 0 0 1
7 241150 67 82.1 0 1 0 90 0 0 1
8 50662 66 84.4 0 1 0 100 0 0 1
9 11987 65 77.2 0 1 0 90 0 0 1
10 140797 65 60.5 0 1 0 90 0 0 1
# … with 2,129 more rows, and 23 more variables: preanti <dbl>,
# race <dbl>, gender <dbl>, str2 <dbl>, strat <dbl>, symptom <dbl>,
# treat <dbl>, offtrt <dbl>, cd40 <dbl>, cd420 <dbl>, cd496 <dbl>,
# r <dbl>, cd80 <dbl>, cd820 <dbl>, cens <dbl>, days <dbl>, arms <dbl>,
# age_m <dbl>, weight_lb <dbl>, cd_change_20 <dbl>, cd_change_960 <dbl>,
# gender_char <chr>, arms_char <chr>- You can sort the rows of dataframes with multiple columns by including many arguments to
arrange(). Now sort the data by arms (arms) and then age_y (age_y). Print the result to make sure it looks right!
trial_act <- trial_act %>%
arrange(XXX, XXX)trial_act <- trial_act %>%
arrange(arms, age_y)
trial_act# A tibble: 2,139 x 33
pidnum age_y weight_kg hemo homo drugs karnof oprior z30 zprior
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 960014 13 48.5 1 0 0 100 0 0 1
2 960031 14 48.3 1 0 0 100 0 0 1
3 990071 14 60 1 0 0 100 0 0 1
4 980042 16 63 1 0 0 100 0 1 1
5 171040 17 51.3 0 0 0 90 0 0 1
6 990026 17 103. 1 0 0 100 0 1 1
7 310234 18 57.3 1 0 0 100 0 1 1
8 940519 18 56.8 1 0 0 100 0 1 1
9 211314 19 50.8 0 0 0 90 0 0 1
10 340767 19 74.8 0 1 0 100 0 0 1
# … with 2,129 more rows, and 23 more variables: preanti <dbl>,
# race <dbl>, gender <dbl>, str2 <dbl>, strat <dbl>, symptom <dbl>,
# treat <dbl>, offtrt <dbl>, cd40 <dbl>, cd420 <dbl>, cd496 <dbl>,
# r <dbl>, cd80 <dbl>, cd820 <dbl>, cens <dbl>, days <dbl>, arms <dbl>,
# age_m <dbl>, weight_lb <dbl>, cd_change_20 <dbl>, cd_change_960 <dbl>,
# gender_char <chr>, arms_char <chr>F - Filter specific rows with filter()
- Using the
filter()function, filter only the rows from males but don’t save the result (Hint:gender_char == "male")
trial_act %>%
filter(XXX == "XXX")trial_act %>%
filter(gender_char == "male")# A tibble: 1,771 x 33
pidnum age_y weight_kg hemo homo drugs karnof oprior z30 zprior
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 960014 13 48.5 1 0 0 100 0 0 1
2 960031 14 48.3 1 0 0 100 0 0 1
3 990071 14 60 1 0 0 100 0 0 1
4 980042 16 63 1 0 0 100 0 1 1
5 990026 17 103. 1 0 0 100 0 1 1
6 940519 18 56.8 1 0 0 100 0 1 1
7 340767 19 74.8 0 1 0 100 0 0 1
8 211007 20 72.7 0 1 0 90 0 1 1
9 261065 20 57.3 0 1 0 100 0 0 1
10 490308 20 72 0 1 0 100 0 0 1
# … with 1,761 more rows, and 23 more variables: preanti <dbl>,
# race <dbl>, gender <dbl>, str2 <dbl>, strat <dbl>, symptom <dbl>,
# treat <dbl>, offtrt <dbl>, cd40 <dbl>, cd420 <dbl>, cd496 <dbl>,
# r <dbl>, cd80 <dbl>, cd820 <dbl>, cens <dbl>, days <dbl>, arms <dbl>,
# age_m <dbl>, weight_lb <dbl>, cd_change_20 <dbl>, cd_change_960 <dbl>,
# gender_char <chr>, arms_char <chr>- Create a new dataframe called
trial_act_malethat only contains rows from males (hint: just assign what you did in the previous question to a new object!). After you create the object, print it to make sure it looks right.
trial_act_male <- trial_act %>%
filter(gender_char == "male")
trial_act_male# A tibble: 1,771 x 33
pidnum age_y weight_kg hemo homo drugs karnof oprior z30 zprior
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 960014 13 48.5 1 0 0 100 0 0 1
2 960031 14 48.3 1 0 0 100 0 0 1
3 990071 14 60 1 0 0 100 0 0 1
4 980042 16 63 1 0 0 100 0 1 1
5 990026 17 103. 1 0 0 100 0 1 1
6 940519 18 56.8 1 0 0 100 0 1 1
7 340767 19 74.8 0 1 0 100 0 0 1
8 211007 20 72.7 0 1 0 90 0 1 1
9 261065 20 57.3 0 1 0 100 0 0 1
10 490308 20 72 0 1 0 100 0 0 1
# … with 1,761 more rows, and 23 more variables: preanti <dbl>,
# race <dbl>, gender <dbl>, str2 <dbl>, strat <dbl>, symptom <dbl>,
# treat <dbl>, offtrt <dbl>, cd40 <dbl>, cd420 <dbl>, cd496 <dbl>,
# r <dbl>, cd80 <dbl>, cd820 <dbl>, cens <dbl>, days <dbl>, arms <dbl>,
# age_m <dbl>, weight_lb <dbl>, cd_change_20 <dbl>, cd_change_960 <dbl>,
# gender_char <chr>, arms_char <chr>- Filter only rows for patients under the age of 60 and save the result to
trial_act_young. After you create it, print the object to make sure it looks right.
trial_act_young <- trial_act %>%
filter(age_y < 60)
trial_act_young# A tibble: 2,110 x 33
pidnum age_y weight_kg hemo homo drugs karnof oprior z30 zprior
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 960014 13 48.5 1 0 0 100 0 0 1
2 960031 14 48.3 1 0 0 100 0 0 1
3 990071 14 60 1 0 0 100 0 0 1
4 980042 16 63 1 0 0 100 0 1 1
5 171040 17 51.3 0 0 0 90 0 0 1
6 990026 17 103. 1 0 0 100 0 1 1
7 310234 18 57.3 1 0 0 100 0 1 1
8 940519 18 56.8 1 0 0 100 0 1 1
9 211314 19 50.8 0 0 0 90 0 0 1
10 340767 19 74.8 0 1 0 100 0 0 1
# … with 2,100 more rows, and 23 more variables: preanti <dbl>,
# race <dbl>, gender <dbl>, str2 <dbl>, strat <dbl>, symptom <dbl>,
# treat <dbl>, offtrt <dbl>, cd40 <dbl>, cd420 <dbl>, cd496 <dbl>,
# r <dbl>, cd80 <dbl>, cd820 <dbl>, cens <dbl>, days <dbl>, arms <dbl>,
# age_m <dbl>, weight_lb <dbl>, cd_change_20 <dbl>, cd_change_960 <dbl>,
# gender_char <chr>, arms_char <chr>- A colleague of yours named Tracy wants a dataframe only containing data from females over the age of 40. Create this dataframe with
filter()and call ittrial_act_tracy. After you create the object, print it to make sure it looks right.
trial_act_tracy <- XXX %>%
filter(XXX > XXX & XXX == XXX)trial_act_tracy <- trial_act %>%
filter(age_y > 40 & gender_char == "female")G - Combine dataframes with left_join()
The trial_act_demo.csv file contains additional (fictional) demographic data about the patients, namely the number of days of exercise they get per week, and their highest level of education. Our goal is to add the demographic information to our trial_act data.
In order to combine the two dataframes, we need to find one ‘key’ column that we can use to match rows. Look at both the
trial_actandtrial_act_demodataframes. Which column can we use as the ‘key’ column?Use the
left_join()function to combine thetrial_actandtrial_act_demodatasets, set thebyargument to the name of the key column that is common in both data sets. Assign the result totrial_act.
trial_act <- trial_act %>%
left_join(XX, by = "XX")trial_act <- trial_act %>%
left_join(trial_act_demo, by = "pidnum")- Print your new
trial_actdataframe. Do you now see the demographic data?
trial_act# A tibble: 2,139 x 35
pidnum age_y weight_kg hemo homo drugs karnof oprior z30 zprior
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 960014 13 48.5 1 0 0 100 0 0 1
2 960031 14 48.3 1 0 0 100 0 0 1
3 990071 14 60 1 0 0 100 0 0 1
4 980042 16 63 1 0 0 100 0 1 1
5 171040 17 51.3 0 0 0 90 0 0 1
6 990026 17 103. 1 0 0 100 0 1 1
7 310234 18 57.3 1 0 0 100 0 1 1
8 940519 18 56.8 1 0 0 100 0 1 1
9 211314 19 50.8 0 0 0 90 0 0 1
10 340767 19 74.8 0 1 0 100 0 0 1
# … with 2,129 more rows, and 25 more variables: preanti <dbl>,
# race <dbl>, gender <dbl>, str2 <dbl>, strat <dbl>, symptom <dbl>,
# treat <dbl>, offtrt <dbl>, cd40 <dbl>, cd420 <dbl>, cd496 <dbl>,
# r <dbl>, cd80 <dbl>, cd820 <dbl>, cens <dbl>, days <dbl>, arms <dbl>,
# age_m <dbl>, weight_lb <dbl>, cd_change_20 <dbl>, cd_change_960 <dbl>,
# gender_char <chr>, arms_char <chr>, exercise <dbl>, education <chr>H - Reshaping with gather() and spread()
Remember the CD4_wide dataframe you created before? Currently it is in the wide format, where each row is a patient, where key data (different CD4 T cell counts) are in different columns like this:
# Data is in a 'wide' format
CD4_wide# A tibble: 2,139 x 5
pidnum arms cd40 cd420 cd496
<dbl> <dbl> <dbl> <dbl> <dbl>
1 960014 0 283 271 NA
2 960031 0 481 428 519
3 990071 0 166 169 28
4 980042 0 299 214 124
5 171040 0 549 415 436
6 990026 0 373 218 NA
7 310234 0 445 371 338
8 940519 0 276 150 34
9 211314 0 298 267 NA
10 340767 0 503 452 NA
# … with 2,129 more rowsOur goal is to convert this data to a ‘long’ format, where each row represents a single CD4 T cell count for a specific patient, like this:
# This is the same data in 'long' format
CD4_long# A tibble: 6,417 x 4
pidnum arms time value
<dbl> <dbl> <chr> <dbl>
1 960014 0 cd40 283
2 960031 0 cd40 481
3 990071 0 cd40 166
4 980042 0 cd40 299
5 171040 0 cd40 549
6 990026 0 cd40 373
7 310234 0 cd40 445
8 940519 0 cd40 276
9 211314 0 cd40 298
10 340767 0 cd40 503
# … with 6,407 more rows- Using the
gather()function, create a new dataframe calledCD4_longthat shows theCD4_widedata in the ‘long’ format. To do this, use the following template. Set the grouping column totimeand the new data column tovalue.
CD4_long <- CD4_wide %>%
gather(XX, # New grouping column
XX, # New data column
-pidnum, -arms) # Names of columns to replicateCD4_long <- CD4_wide %>%
gather(time, # New grouping column
value, # New data column
-pidnum, -arms) # Names of columns to replicate- Print your
CD4_longdataframe! Do you now see that each row is a specific observation for a patient?
CD4_long# A tibble: 6,417 x 4
pidnum arms time value
<dbl> <dbl> <chr> <dbl>
1 960014 0 cd40 283
2 960031 0 cd40 481
3 990071 0 cd40 166
4 980042 0 cd40 299
5 171040 0 cd40 549
6 990026 0 cd40 373
7 310234 0 cd40 445
8 940519 0 cd40 276
9 211314 0 cd40 298
10 340767 0 cd40 503
# … with 6,407 more rows- Now use the
spread()function to convert the long data bring the data back into the wide format! To do this, make the first argumenttime, and the second argumentvalue(you don’t need to save the object)
CD4_long %>%
spread(XX, # Grouping column
XX) # Value columnnCD4_long %>%
spread(time, value)# A tibble: 2,139 x 5
pidnum arms cd40 cd420 cd496
<dbl> <dbl> <dbl> <dbl> <dbl>
1 10056 2 422 477 660
2 10059 3 162 218 NA
3 10089 3 326 274 122
4 10093 3 287 394 NA
5 10124 0 504 353 660
6 10140 1 235 339 264
7 10165 0 244 225 106
8 10190 0 401 366 453
9 10198 3 214 107 8
10 10229 0 221 132 NA
# … with 2,129 more rowsI - Practice
- Create a new dataframe called
trial_Afromtrial_actwith the following restrictions:
- Only patients who take intravenous drugs (
drugs == 1) and… - Only patients younger than 60 and…
- Only patients whose baseline CD4 T cell counts
cd40were greater than 250 and less than 400.
trial_A <- trial_act %>%
filter(drugs == 1,
age_y < 60,
cd40 > 250 & cd40 < 400)
trial_A# A tibble: 126 x 35
pidnum age_y weight_kg hemo homo drugs karnof oprior z30 zprior
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 630023 26 86.2 0 0 1 90 0 0 1
2 10378 27 67.3 0 0 1 90 0 1 1
3 320475 27 66.6 0 0 1 90 0 0 1
4 71333 29 73.9 0 1 1 100 0 0 1
5 140809 29 70 0 1 1 100 0 0 1
6 630030 29 88.2 0 0 1 100 0 0 1
7 190395 33 68 0 0 1 100 0 1 1
8 320365 34 86.0 0 0 1 100 0 0 1
9 540015 34 72.6 0 0 1 100 0 0 1
10 180917 35 77.7 0 0 1 90 0 1 1
# … with 116 more rows, and 25 more variables: preanti <dbl>, race <dbl>,
# gender <dbl>, str2 <dbl>, strat <dbl>, symptom <dbl>, treat <dbl>,
# offtrt <dbl>, cd40 <dbl>, cd420 <dbl>, cd496 <dbl>, r <dbl>,
# cd80 <dbl>, cd820 <dbl>, cens <dbl>, days <dbl>, arms <dbl>,
# age_m <dbl>, weight_lb <dbl>, cd_change_20 <dbl>, cd_change_960 <dbl>,
# gender_char <chr>, arms_char <chr>, exercise <dbl>, education <chr>- Add a new column to
trial_actcalleddrugs_charwhich is"User"when drugs == 1, and"Non User"when drugs == 0.
trial_act <- trial_act %>%
mutate(drugs_char = case_when(
drugs == 1 ~ "User",
drugs == 0 ~ "Non User"
))
trial_act# A tibble: 2,139 x 36
pidnum age_y weight_kg hemo homo drugs karnof oprior z30 zprior
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 960014 13 48.5 1 0 0 100 0 0 1
2 960031 14 48.3 1 0 0 100 0 0 1
3 990071 14 60 1 0 0 100 0 0 1
4 980042 16 63 1 0 0 100 0 1 1
5 171040 17 51.3 0 0 0 90 0 0 1
6 990026 17 103. 1 0 0 100 0 1 1
7 310234 18 57.3 1 0 0 100 0 1 1
8 940519 18 56.8 1 0 0 100 0 1 1
9 211314 19 50.8 0 0 0 90 0 0 1
10 340767 19 74.8 0 1 0 100 0 0 1
# … with 2,129 more rows, and 26 more variables: preanti <dbl>,
# race <dbl>, gender <dbl>, str2 <dbl>, strat <dbl>, symptom <dbl>,
# treat <dbl>, offtrt <dbl>, cd40 <dbl>, cd420 <dbl>, cd496 <dbl>,
# r <dbl>, cd80 <dbl>, cd820 <dbl>, cens <dbl>, days <dbl>, arms <dbl>,
# age_m <dbl>, weight_lb <dbl>, cd_change_20 <dbl>, cd_change_960 <dbl>,
# gender_char <chr>, arms_char <chr>, exercise <dbl>, education <chr>,
# drugs_char <chr>- Create a new dataframe called
trial_Bfromtrial_actwith the following restrictions:
- Only patients whose CD4 T cell counts increase more than 200 between baseline (
cd40) and after 96 weeks (cd496) - Only drug users
trial_B <- trial_act %>%
filter(
cd496 - cd40 > 200,
drugs == 1
)
trial_B# A tibble: 8 x 36
pidnum age_y weight_kg hemo homo drugs karnof oprior z30 zprior
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 220489 45 70.9 0 0 1 100 0 0 1
2 140744 30 81.2 0 1 1 100 0 0 1
3 10962 41 81.9 0 0 1 90 0 0 1
4 50580 68 90.5 0 1 1 100 0 1 1
5 10881 36 81.6 0 1 1 100 0 1 1
6 250197 49 86.2 0 1 1 90 0 0 1
7 50572 28 84.4 0 1 1 90 0 0 1
8 50623 35 77 0 1 1 100 0 0 1
# … with 26 more variables: preanti <dbl>, race <dbl>, gender <dbl>,
# str2 <dbl>, strat <dbl>, symptom <dbl>, treat <dbl>, offtrt <dbl>,
# cd40 <dbl>, cd420 <dbl>, cd496 <dbl>, r <dbl>, cd80 <dbl>,
# cd820 <dbl>, cens <dbl>, days <dbl>, arms <dbl>, age_m <dbl>,
# weight_lb <dbl>, cd_change_20 <dbl>, cd_change_960 <dbl>,
# gender_char <chr>, arms_char <chr>, exercise <dbl>, education <chr>,
# drugs_char <chr>X - Advanced
- Read the
appointments.csvand theweather.csvfiles into two objects calledappointmentsandweather, respectively, using the code below. Theappointmentsdataset contains data on medical appointment no shows in Brazil including a set of other variables. Theweatherdataset contains data on the weather of the location (same for all cases) of the appointment.
# read new data sets
appointments <- read_csv('1_Data/appointments.csv')
weather <- read_csv('1_Data/weather.csv')- Your goal is to use these datasets to answer the question: “Are patients less likely to show up for their appointments on rainy days than sunny days?” To do this, consider the following steps
- Explore each dataset visually by printing it and/or using
View() - Look at the variable descriptions in the Datasets tab above to understand what the variables mean.
- Think about which columns in each dataset are necessary to answer your question (e.g.; how do you define rainy versus non-rainy days? Which variable(s) indicate whether a patient showed up for their appointment or not?).
- Using
mutate()andcase_when(), add a new column to theweatherdata calledRainywhich isTRUEon a rainy day, andFALSEon a non-rainy day. - Combine (aka, join) the two datasets into a dataset called
combinedbased on a key column usingleft_join() - Using
filter(), create a dataset calledrainy_appointmentscontaining only appointment data for rainy days. - Using
filter(), create a dataset calleddry_appointmentscontaining only appointment data for dry days. - For the
rainy_appointmentsdata, calculate the perentage of patients who did not show up for their appointment. - For the
dry_appointmentsdata, calculate the same percentage.
# read new data sets
appointments <- read_csv('1_Data/appointments.csv')
weather <- read_csv('1_Data/weather.csv')
# Change name of weather YYYMMDD to DAY
weather <- weather %>%
rename(DAY = YYYYMMDD)
# Define badf days as PRECTOT > 1
weather <- weather %>%
mutate(Rainy = case_when(PRECTOT > 1 ~ TRUE,
PRECTOT <= 1 ~ FALSE))
# Change name of appointments date
appointments <- appointments %>%
rename(DAY = AppointmentDay)
# Join weather to appointments
appointments <- appointments %>%
left_join(weather, by = "DAY")
# Define wet_day_appointments
rainy_appointments <- appointments %>%
filter(Rainy == TRUE)
# Define sunny days
dry_appointments <- appointments %>%
filter(Rainy == FALSE)
# What percent of people don't show up on rainy days?
mean(rainy_appointments$NoShow == "Yes")[1] 0.207# What percent of people don't show up on dry days?
mean(dry_appointments$NoShow == "Yes")[1] 0.2# Answer: I don't find much of a difference! What about you?Examples
# Wrangling with dplyr and tidyr ---------------------------
library(tidyverse) # Load tidyverse for dplyr and tidyr
# Load baselers data
baselers <- read_csv("https://raw.githubusercontent.com/therbootcamp/baselers/master/inst/extdata/baselers.txt")
# Perform many dplyr operations in a row
baselers %>%
# Change some names
rename(age_y = age,
swimming = rhine) %>%
# Only include people over 30
filter(age_y > 30) %>%
# Calculate some new columns
mutate(weight_lbs = weight * 2.22,
height_m = height / 100,
BMI = weight / height_m ^ 2,
# Make binary version of sex
sex_bin = case_when(
sex == "male" ~ 0,
sex == "female" ~ 1),
# Show when height is greater than 150
height_lt_150 = case_when(
height < 150 ~ 1,
height >= 150 ~ 0)) %>%
# Sort in ascending order of sex, then
# descending order of age
arrange(sex, desc(age_y)))
# TIPP -------
# In this practical you will do many operations on dataframes. Remember: when using `dplyr`, you can chain
# multiple functions together with the pipe `%>%`. When giving your answers to the questions in this practical,
# see how many operations you can chain with the pipe!
baselers <- read_csv("1_Data/baselers.csv")
# Method 1: Separate operations
baselers <- baselers %>%
rename(Age_y = age) # Change age to Age__y
baselers <- baselers %>%
mutate(food_p = food / income) # calculate food_p
baselers <- baselers %>%
filter(sex == "m") # Only include males
# Method 2: Chain with the pipe!
baselers <- read_csv("1_Data/baselers.csv")
baselers <- baselers %>%
rename(Age_y = age) %>% # Change age to Age__y
mutate(food_p = food / income) %>% # calculate food_p
filter(sex == "m") # Only include malesDatasets
| File | Rows | Columns |
|---|---|---|
| trial_act.csv | 2139 | 27 |
| trial_act_demo_fake.csv | 2139 | 3 |
| appointments.csv | 110526 | 13 |
| weather.csv | 41 | 10 |
trial_act.csv
First 5 rows of trial_act.csv
| pidnum | age | wtkg | hemo | homo | drugs | karnof | oprior | z30 | zprior | preanti | race | gender | str2 | strat | symptom | treat | offtrt | cd40 | cd420 | cd496 | r | cd80 | cd820 | cens | days | arms |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10056 | 48 | 89.8 | 0 | 0 | 0 | 100 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 422 | 477 | 660 | 1 | 566 | 324 | 0 | 948 | 2 |
| 10059 | 61 | 49.4 | 0 | 0 | 0 | 90 | 0 | 1 | 1 | 895 | 0 | 0 | 1 | 3 | 0 | 1 | 0 | 162 | 218 | NA | 0 | 392 | 564 | 1 | 1002 | 3 |
| 10089 | 45 | 88.5 | 0 | 1 | 1 | 90 | 0 | 1 | 1 | 707 | 0 | 1 | 1 | 3 | 0 | 1 | 1 | 326 | 274 | 122 | 1 | 2063 | 1893 | 0 | 961 | 3 |
| 10093 | 47 | 85.3 | 0 | 1 | 0 | 100 | 0 | 1 | 1 | 1399 | 0 | 1 | 1 | 3 | 0 | 1 | 0 | 287 | 394 | NA | 0 | 1590 | 966 | 0 | 1166 | 3 |
| 10124 | 43 | 66.7 | 0 | 1 | 0 | 100 | 0 | 1 | 1 | 1352 | 0 | 1 | 1 | 3 | 0 | 0 | 0 | 504 | 353 | 660 | 1 | 870 | 782 | 0 | 1090 | 0 |
The trial_act.csv data set contains a randomized clinical trial to compare monotherapy with zidovudine or didanosine with combination therapy with zidovudine and didanosine or zidovudine and zalcitabine in adults infected with the human immunodeficiency virus type I whose CD4 T cell counts were between 200 and 500 per cubic millimeter.
| Name | Description |
|---|---|
| pidnum | patient’s ID number |
| age | age in years at baseline |
| wtkg | weight in kg at baseline |
| hemo | hemophilia (0=no, 1=yes) |
| homo | homosexual activity (0=no, 1=yes) |
| drugs | history of intravenous drug use (0=no, 1=yes) |
| karnof | Karnofsky score (on a scale of 0-100) |
| oprior | non-zidovudine antiretroviral therapy prior to initiation of study treatment (0=no, 1=yes) |
| z30 | zidovudine use in the 30 days prior to treatment initiation (0=no, 1=yes) |
| zprior | zidovudine use prior to treatment initiation (0=no, 1=yes) |
| preanti | number of days of previously received antiretroviral therapy |
| race | race (0=white, 1=non-white) |
| gender | gender (0=female, 1=male) |
| str2 | antiretroviral history (0=naive, 1=experienced) |
| strat | antiretroviral history stratification (1=’antiretroviral naive’, 2=’> 1 but ≤ 52 weeks of prior antiretroviral therapy’, 3=’> 52 weeks’) |
| symptom | symptomatic indicator (0=asymptomatic, 1=symptomatic) |
| treat | treatment indicator (0=zidovudine only, 1=other therapies) |
| offtrt | indicator of off-treatment before 96±5 weeks (0=no,1=yes) |
| cd40 | CD4 T cell count at baseline |
| cd420 | CD4 T cell count at 20±5 weeks |
| cd496 | CD4 T cell count at 96±5 weeks (=NA if missing) |
| r | missing CD4 T cell count at 96±5 weeks (0=missing, 1=observed) |
| cd80 | CD8 T cell count at baseline |
| cd820 | CD8 T cell count at 20±5 weeks |
| cens | indicator of observing the event in days |
| days | number of days until the first occurrence of: (i) a decline in CD4 T cell count of at least 50 (ii) an event indicating progression to AIDS, or (iii) death. |
| arms | treatment arm (0=zidovudine, 1=zidovudine and didanosine, 2=zidovudine and zalcitabine, 3=didanosine). |
trial_act_demo_fake.csv
First 5 rows of trial_act_demo_fake.csv
| pidnum | exercise | education |
|---|---|---|
| 10931 | 4 | HS |
| 11971 | 1 | <HS |
| 330244 | 1 | HS |
| 270879 | 1 | BA |
| 11435 | 0 | <HS |
The trial_act_demo_fake.csv data set contains fake demogrpahic information corresponding to the patients in trial_act.csv
| Name | Description |
|---|---|
| pidnum | patient’s ID number |
| exercise | the number of days per week that the patient exercises |
| education | the patient’s education level |
appointments.csv
First 5 rows of appointments.csv
| PatientId | AppointmentID | Gender | ScheduledDay | AppointmentDay | Age | Neighbourhood | Hypertension | Diabetes | Alcoholism | Handicap | SMS_received | NoShow |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.99e+13 | 5642903 | F | 2016-04-29 18:38:08 | 2016-04-29 | 62 | JARDIM DA PENHA | TRUE | FALSE | FALSE | 0 | FALSE | No |
| 5.59e+14 | 5642503 | M | 2016-04-29 16:08:27 | 2016-04-29 | 56 | JARDIM DA PENHA | FALSE | FALSE | FALSE | 0 | FALSE | No |
| 4.26e+12 | 5642549 | F | 2016-04-29 16:19:04 | 2016-04-29 | 62 | MATA DA PRAIA | FALSE | FALSE | FALSE | 0 | FALSE | No |
| 8.68e+11 | 5642828 | F | 2016-04-29 17:29:31 | 2016-04-29 | 8 | PONTAL DE CAMBURI | FALSE | FALSE | FALSE | 0 | FALSE | No |
| 8.84e+12 | 5642494 | F | 2016-04-29 16:07:23 | 2016-04-29 | 56 | JARDIM DA PENHA | TRUE | TRUE | FALSE | 0 | FALSE | No |
| Variable | Description |
|---|---|
| PatientId | ID of a patient |
| AppointmentID | ID for each appointment |
| Gender | Male or Female |
| ScheduledDay | The day of the actual appointment, when patients have to visit the doctor |
| AppointmentDay | The day someone called or registered the appointment, this should be before the appointment |
| Age | How old is the patient |
| Neighbourhood | Where the patient was born |
| Hipertension | True or False |
| Diabetes | True or False |
| Alcoholism | True or False |
| Handcap | Hanicapped - level 1:4, 1 lowest level |
| SMS_received | True: 1 or more messages sent to the patient |
| No-show | 1: No, 2: Yes |
weather.csv
First 5 rows of weather.csv
| LON | LAT | YEAR | MM | DD | DOY | YYYYMMDD | RH2M | T2M | PRECTOT |
|---|---|---|---|---|---|---|---|---|---|
| 20.3 | 40.3 | 2016 | 4 | 29 | 120 | 2016-04-29 | 86.6 | 11.57 | 6.18 |
| 20.3 | 40.3 | 2016 | 4 | 30 | 121 | 2016-04-30 | 73.5 | 14.02 | 0.04 |
| 20.3 | 40.3 | 2016 | 5 | 1 | 122 | 2016-05-01 | 75.9 | 12.98 | 4.48 |
| 20.3 | 40.3 | 2016 | 5 | 2 | 123 | 2016-05-02 | 84.7 | 11.41 | 22.99 |
| 20.3 | 40.3 | 2016 | 5 | 3 | 124 | 2016-05-03 | 84.6 | 8.69 | 5.81 |
| Variable | Description |
|---|---|
| LON | Longitude |
| LAT | Latitude |
| YEAR | Year |
| MM | Month |
| DD | Day |
| DOY | Day of Year |
| YYYYMMDD | Date |
| RH2M | Relative Humidity at 2 Meters |
| T2M | Temperature at 2 Meters |
| PRECTOT | Precipitation |
Functions
Packages
| Package | Installation |
|---|---|
tidyverse |
install.packages("tidyverse") |
Functions
| Function | Package | Description |
|---|---|---|
rename() |
dplyr |
Rename columns |
select() |
dplyr |
Select columns based on name or index |
filter() |
dplyr |
Select rows based on some logical criteria |
arrange() |
dplyr |
Sort rows |
mutate() |
dplyr |
Add new columns |
case_when() |
dplyr |
Recode values of a column |
group_by(), summarise() |
dplyr |
Group data and then calculate summary statistics |
left_join() |
dplyr |
Combine multiple data sets using a key column |
spread() |
tidyr |
Convert long data to wide format - from rows to columns |
gather() |
tidyr |
Convert wide data to long format - from columns to rows |
Resources
dplyr vignette
See https://cran.r-project.org/web/packages/dplyr/vignettes/dplyr.html for the full dplyr vignette with lots of wrangling tips and tricks.
Cheatsheets
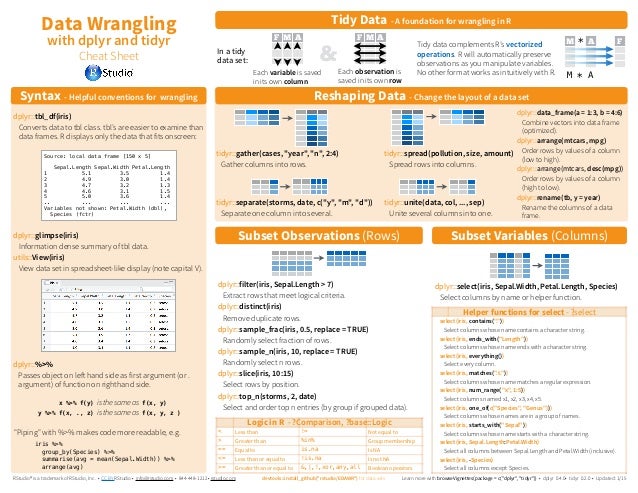
from R Studio
